MIIDL /ˈmaɪdəl/ is a Python package for microbial biomarkers identification powered by interpretable deep learning.
Getting Started
👋Welcome!
🔗This guide will provide you with a specific example that using miidl to detect microbial biomarkers of colorectal cancer and predict clinical outcomes.
After that, you will learn how to use this tool properly.
Installation
pip install miidl
or
conda install miidl captum -c pytorch -c conda-forge -c bioconda
Features
- One-stop profiling
- Multiple strategies for biological data
- More interpretable, not a “black box”
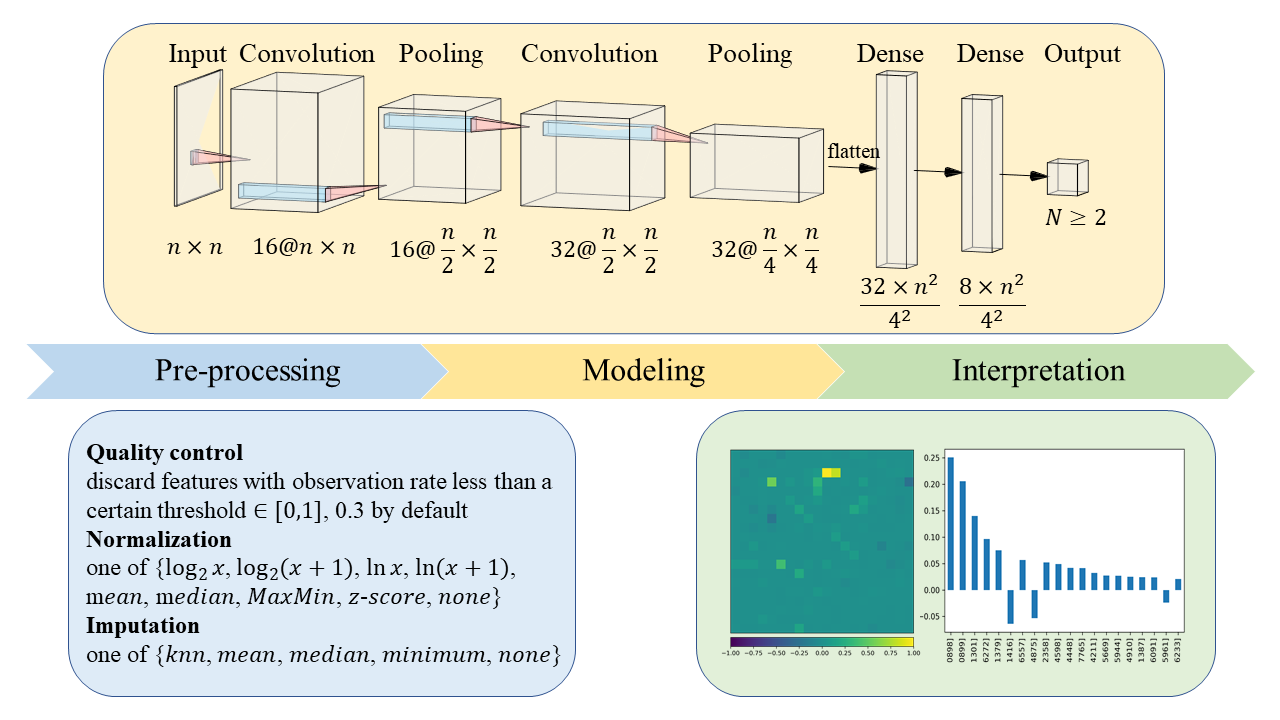
Workflow
1) Quality Control
The very first procedure is filtering features according to a threshold of observation (non-missing) rate (0.3 by default).
2) Normalization
miidl offers plenty of normalization methods to transform data and make samples more comparable.
3) Imputation
By default, this step is unactivated, as miidl is designed to solve problems including sparseness. But imputation can be useful in some cases. Commonly used methods are available if needed.
4) Reshape
The pre-processed data also need to be zero-completed to a certain length, so that a CNN model can be applied.
5) Modeling
A CNN classifier is trained for discrimination. PyTorch is needed.
6) Interpretation
Captum is dedicated to model interpretability for PyTorch. This step relies heavily on captum.
Contact
If you have further thoughts or queries, please feel free to email at chunribu@mail.sdu.edu.cn or open an issue!
Citation
@misc{jiang2021miidl,
title={MIIDL: a Python package for microbial biomarkers identification powered by interpretable deep learning},
author={Jian Jiang},
year={2021},
eprint={2109.12204},
archivePrefix={arXiv},
primaryClass={q-bio.QM}
}
License
MIIDL is released under the MIT license.